-

-
Find Courses
Collections
Cross-Disciplinary Topic Lists
- About
- Donate
- Featured Sites
This is an archived course. A more recent version may be available at ocw.mit.edu.
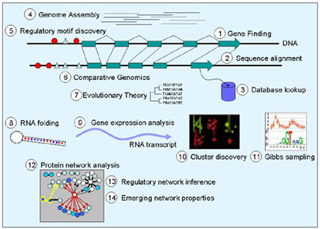
Pictographic representation of the challenges in computational biology. (Image by Prof. Manolis Kellis.)
Prof. Manolis Kellis
Prof. James Galagan
6.047 / 6.878
Fall 2008
Undergraduate
This course focuses on the algorithmic and machine learning foundations of computational biology, combining theory with practice. We study the principles of algorithm design for biological datasets, and analyze influential problems and techniques. We use these to analyze real datasets from large-scale studies in genomics and proteomics. The topics covered include:
OCW has published multiple versions of this subject. ![]()
Archived versions: ![]()
Manolis Kellis, and James Galagan. 6.047 Computational Biology: Genomes, Networks, Evolution. Fall 2008. Massachusetts Institute of Technology: MIT OpenCourseWare, https://ocw.mit.edu. License: Creative Commons BY-NC-SA.
For more information about using these materials and the Creative Commons license, see our Terms of Use.
